EVLncRNAs2.0 User Tutorial
The usage of the EVLncRNAs database is as follows:
1. In ‘Browse’ menu, there are eight pull-down menus: Species, Disease, Interaction, Structure, Peptide-coding, CircRNA, Exosomal lncRNAs, Resistant lncRNAs. Users can browse relevant entries in EVLncRNAs by clicking any pull-down menu and the page will move to the related page. On each page, there are drop-down boxes to choose from and browse the corresponding categories. Press the ‘Detail’ could jump to the corresponding detailed pageof the lncRNA (Figure 1).
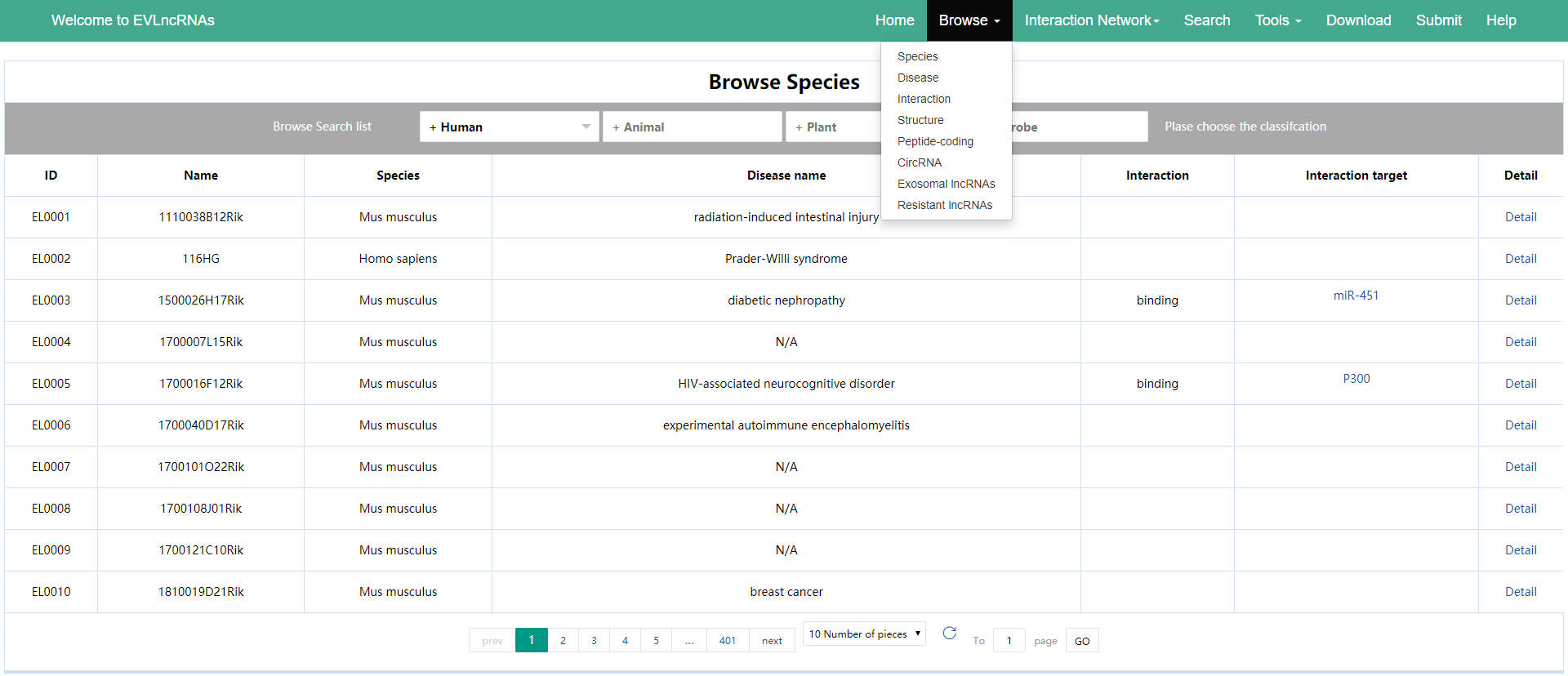
Figure 1. The Browse page of the EVLncRNAs
1) Each entry in ‘Detail’ has the following items (The blank part is not shown):
-
LncRNA Information:
ID, Name, Aliases, Species, Chromosome, Start site, End site, Chain, Extron NO., Type, NCBI accession, Ensembl, Peptide-related, CircRNA, Exosomal, Structure
-
Peptide Information:
Peptide name, length, sequence
-
Structure Information:
Structure view, PDB ID
-
Disease:
Disease, Drug/chemo/stress, Method, Sample, Expression pattern, Dysfunction type, Description, PMID, Source
-
Function:
Drug/chemo/stress, Method, Sample/Condition, Expression pattern, Dysfunction type, Description, PMID, Source
-
Interaction:
Interaction target, Level of interaction, Type of interaction, Description, PMID, Source
2) Function type: According to the experimentally supported associations between lncRNAs and diseases, the function types were classified into expression, mutation, interaction, locus, and epigenetics.
3) Level of interaction: levels of interaction components, for example, RNA-DNA, RNA-RNA, RNA-TF and RNA-Protein.
4) Type of interaction: According to the way how the molecules interact, types of interaction were classified into binding, regulation and co-expression.
2. The comprehensive lncRNA interaction network was constructed to facilitate users to track the underlying relationships among lncRNAs, miRNAs, proteins, genes and other functional elements (Figure 2).

Figure 2 The comprehensive lncRNA interaction network
Notice:
1) The total interaction network includes interaction type of binding, regulation and co-expression.
2) Binding networks are those lncRNAs having direct contact interactions with other functional elements.
3) Users can set their preferred displayed items according to the options on the left. Through clicking the items in the upper left corner of the page, users could choose which items to display. For example, Figure 3 shows the interaction network of only lncRNAs and diseases.

Figure 3 The lncRNA and disease interaction network
4) Use the mouse wheel to zoom in and out of the network.
5) Hold down the left mouse button for more than two seconds to drag the network.
6) When an item is selected with the mouse, its associated factors and diseases are highlighted (Figure 4).
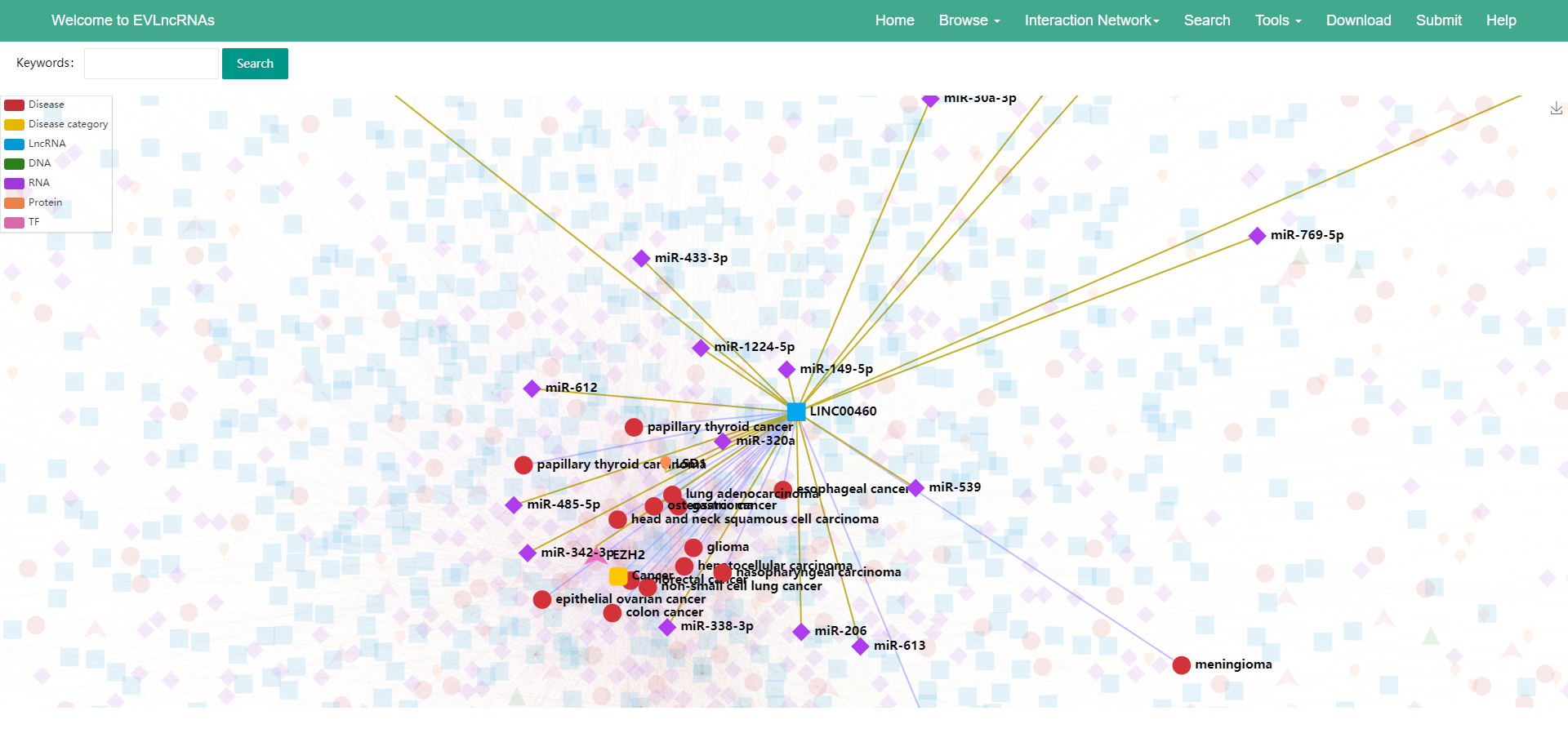
Figure 4 The selected item and its associated factors and diseases are highlighted.
7) The relations between lncRNAs and diseases and between lncRNAs and biomolecules are distinguished in purple and yellow colors, respectively (Figure 4).
8) Using the search box in the upper left corner, users can search for the lncRNA or interaction target of interest, and then the small interaction network of the search keyword will be displayed (Figure 5).
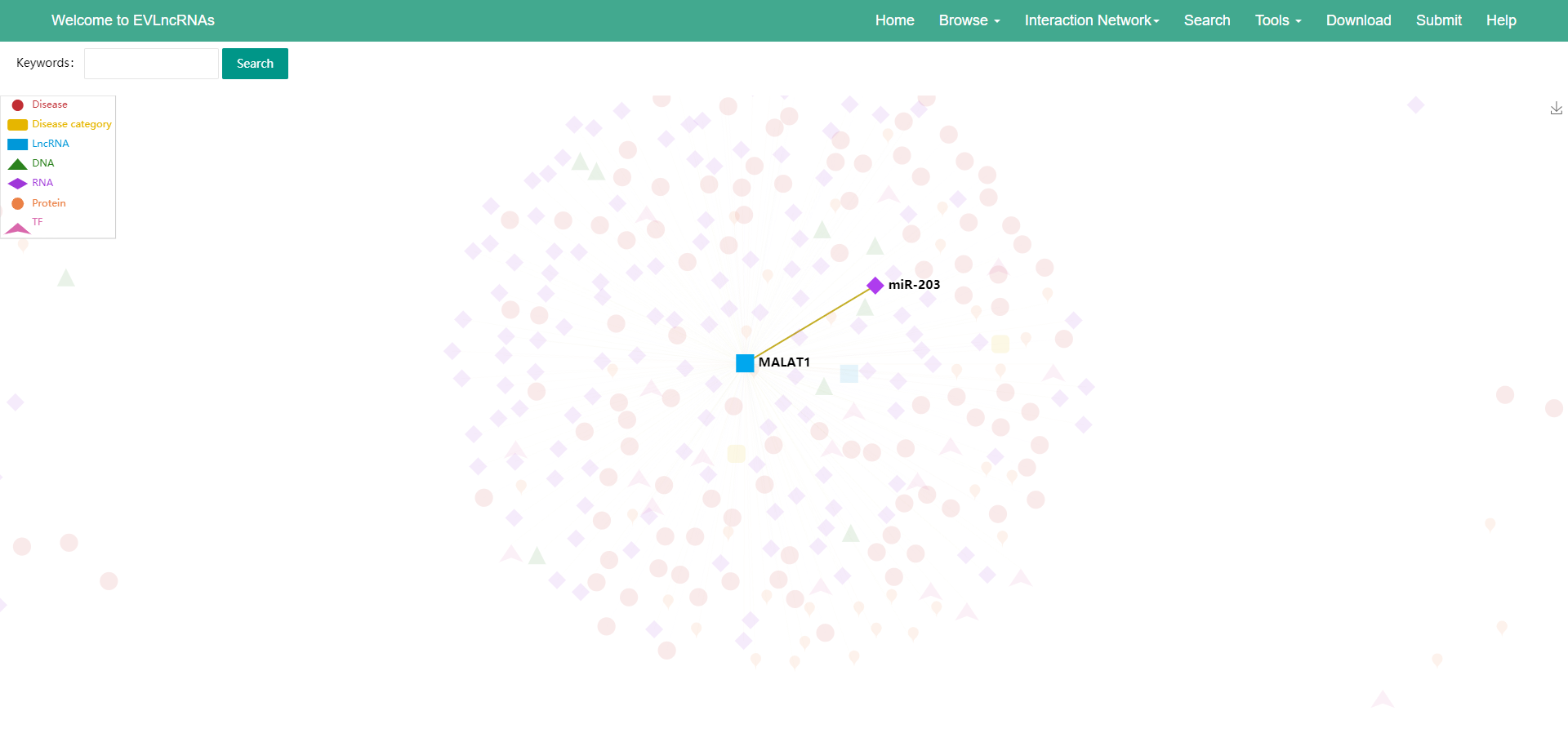
Figure 5 The search result of lncRNA MALAT1.
3. In the ‘Search’ page, EVLncRNAs enables users to search by lncRNA name, disease name, associated components, and so on. EVLncRNAs offers ‘fuzzy’ searching capabilities and returns all matching records. The search results are linked to the detailed pages of related lncRNAs (Figure 6).

Figure 6 The result page of search
4. EVLncRNAs provides reliable and available tools for lncRNA prediction and lncRNA function prediction in the ‘Tools’ page.
5. EVLncRNAs supports users to download the database data in the ‘Download’ page.
6. EVLncRNAs invites users to submit novel experimentally validated lncRNAs and related diseases or associated components in ‘Submit’ page. The submitted record approved by our researchers will be included in the following release.
If there are any comments or suggestions, please contact at evlncrnas@163.com